To decipher the functional impact of proteoforms, most research projects within the SNP2Prot will focus on proteins whose biochemical functions and biological context are reasonably well understood. Each project will apply established biochemical or molecular assays to detect functional differences between the selected proteoforms. To ensure maximum benefits of the available and complementary expertise within this consortium, all research projects will be headed by two PIs from the corresponding fields of plant and protein sciences (or bioinformatics).
For several projects and their associated questions, it will suffice to uncover functional consequences of different proteoforms on various molecular processes (localisation, modification, stability, binding of nucleic acids or other proteins, enzymatic reaction products, etc.) or phenotypic properties (growth, development). However, the aspired ‘gold standard’ will always be investigating the effect of an nsSNP on protein folding and 3-D structure by biophysical methods (e.g., XL-MS, NMR, crystallisation, or cryo-EM).
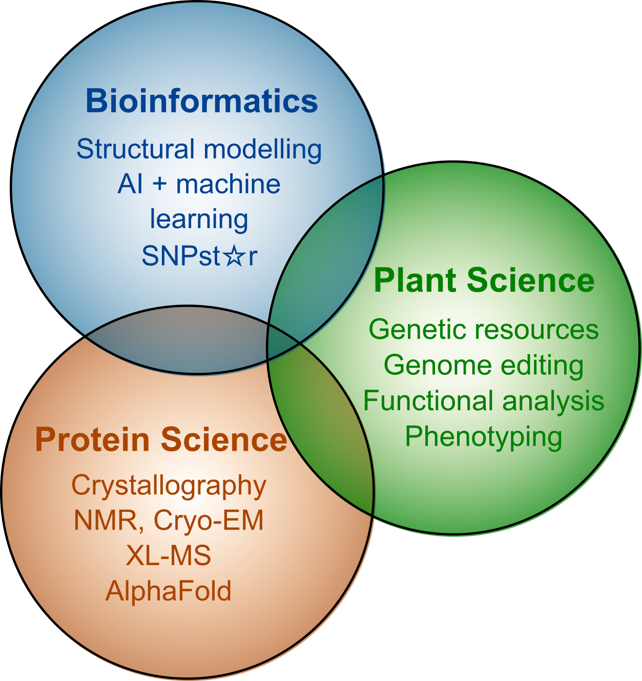
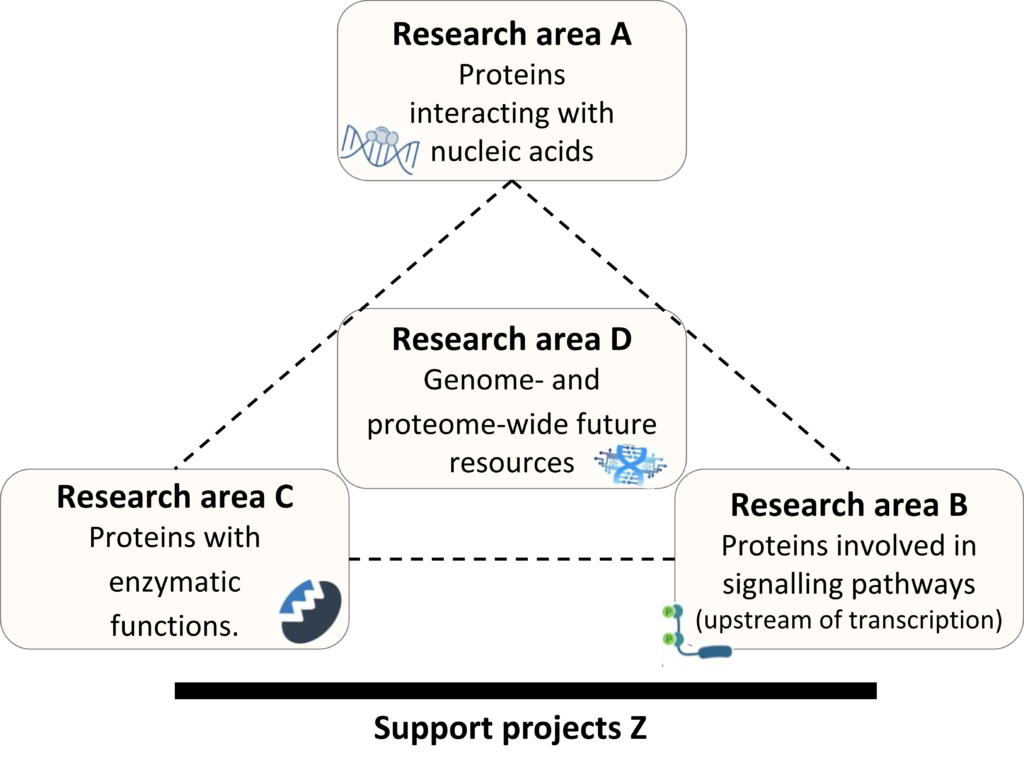
The projects of SN2Prot analyse different functional classes of proteins which are assorted into research areas A, B and C. Research area D serves as an integrative research area and generates valuable resources to open new avenues for the whole consortium. Z-pojects support all SNP2Prot projects in terms of experimental research (Z02) and central administration (Z01).
Research area A // Proteins interacting with nucleid acids
Research area B // Proteins involved in signalling pathways (upstream of transcription)
Research area C // Proteins with enzymatic functions
Research area D // Genome- and proteome-wide future resources
