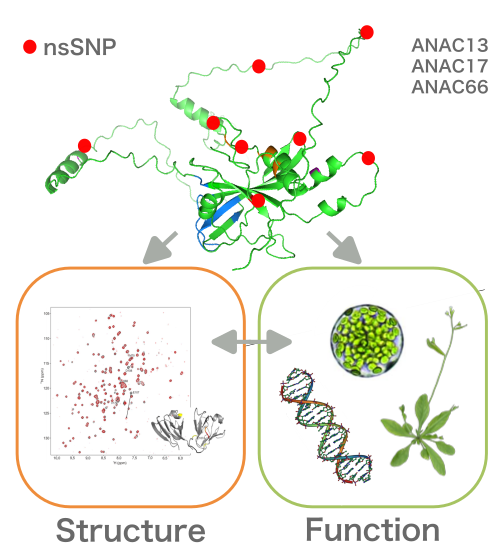
Structure-function analysis of NAC transcription factor proteoforms
NAC transcription factors (TFs) regulate diverse processes ranging from meristem identity and secondary wall synthesis to numerous stress responses. Canonical NAC proteins contain a highly conserved N-terminal NAC domain and a diverse, intrinsically disordered C-terminal region. DNA binding and TF dimerization are generally mediated via the conserved NAC domain whereas the unstructured C-terminal domain is required for transcriptional regulation of the respective target genes and may contribute to target specificity. Structural information available so far is limited to the NAC domain which allowed the identification of critical amino acid residues but so far fails to explain the highly divers function of different NACs.
This project will combine biophysical NMR and molecular biological analyses to provide a novel structure-function analysis of NAC TFs. We will exploit naturally occurring non-synonymous single nucleotide polymorphism (nsSNP) within the Arabidopsis gene pool to: (i) identify amino acid residues within the NAC domain that affect NAC domain structure and protein, (ii) assess the impact of naturally occurring variation within the intrinsically disordered region on the capacity for transcriptional regulation, NAC domain structure and function, and (iii) to assess the biological relevance of individual proteoform function by in vivo analyses.
Project related publications:
Zhu, Z., Trenner, J., Delker, C., & Quint, M. (2024). Tracing the Evolutionary History of the Temperature-Sensing Prion-like Domain in EARLY FLOWERING 3 Highlights the Uniqueness of AtELF3. Molecular Biology and Evolution, 41(10). https://doi.org/10.1093/molbev/msae205


