Impact of natural allelic variation on structure and function of the immune proteases SAP1 and SAP2
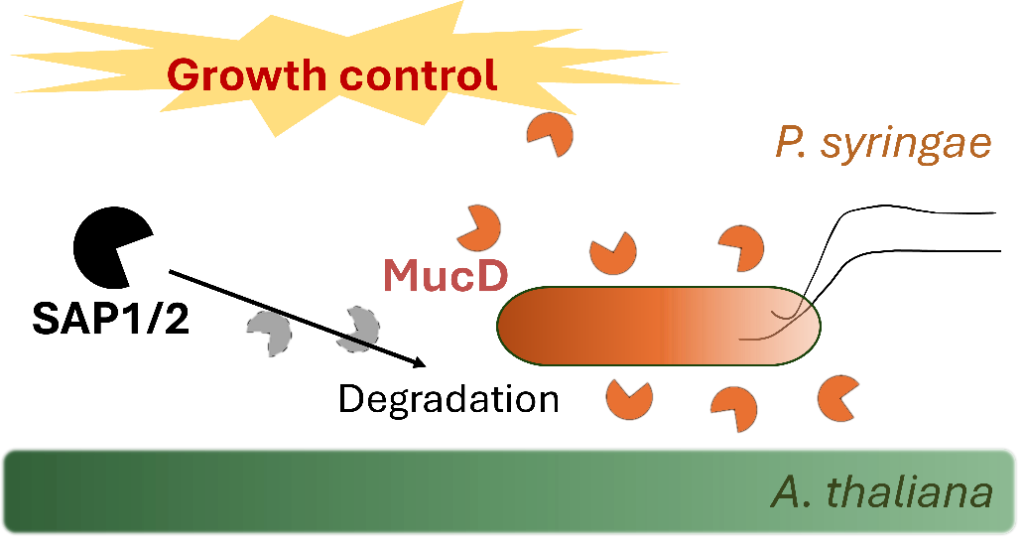
Plant diseases caused by pathogens pose significant threats to ecosystem health and crop productivity globally. Plants have evolved an intricate immune system to combat these pathogens, including mechanisms to directly suppress their growth. Engineering plant responses offers a sustainable solution for crop protection, with direct growth suppression mechanisms being particularly promising due to minimal impact on plant growth. Arabidopsis secreted aspartic proteases (SAP1 and SAP2) have been identified as effective inhibitors of the bacterial pathogen Pseudomonas syringae by cleaving the conserved bacterial protein MucD. These proteases are conserved across plant species and are promising targets for protease engineering in crop protection. This project aims to understand how discrete amino acid changes affect protease function and structure by studying natural variations in Arabidopsis SAP genes. Methods include heterologous expression, purification, and characterization of different proteoforms, as well as assessing their efficacy in bacterial growth suppression and substrate specificity. Structural analysis based on structural mass spectrometry (cross-linking MS, hydrogen-deuterium exchange and native MS) and computational modeling will provide insights into structure-function relationships and guide rational design of these proteases for crop protection.
Project related publications:
Schuster, M., Ciattoni, A. L., & van der Hoorn, R. A. L. (2025). Hacking the immune system: Plant immune protease engineering for crop protection. Journal of Experimental Botany. Advance online publication. https://doi.org/10.1093/jxb/eraf137


