Functional proteoform variation in thermomorphogenesis master regulators
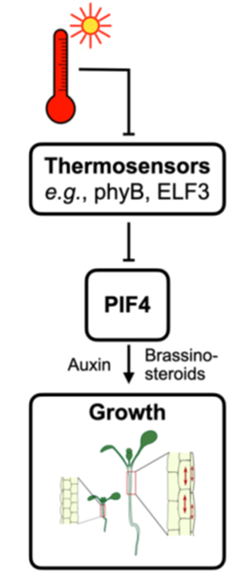
Morphological changes in response to temperature are a specific type of phenotypic plasticity called thermomorphogenesis. The canonical thermomorphogenesis signalling pathway revolves around the major hub and transcription factor (TF) PHYTOCHROME-INTERACTING FACTOR 4 (PIF4). At low temperatures, PIF4 is negatively regulated by thermosensors like PHYTOCHROME B (phyB) or EARLY FLOWERING 3 (ELF3). At high temperatures, PIF4 is derepressed and induces the expression of growth-promoting genes. Associations between natural alleles and phenotypic growth responses have been shown for all three genes by others as well as by us. Despite these associations, the functional variation between proteoforms remains not at all understood. The biological question of A02 therefore focuses on a mechanistic understanding of functional differences between PIF4 and ELF3 proteoforms primarily in thermomorphogenesis, but also in related biological processes like input and output of the circadian clock.
We will focus on DNA binding, protein-protein interactions, and the generation of a structural understanding of these intrinsically disordered proteins. On various levels, cross-linking mass spectrometry (XL-MS), which has emerged as a critical stride forward in elucidating molecular interactions and structural dynamics of proteins, will be our primary methodological approach.
Project related publications:
Zhu, Z., Trenner, J., Delker, C., & Quint, M. (2024). Tracing the Evolutionary History of the Temperature-Sensing Prion-like Domain in EARLY FLOWERING 3 Highlights the Uniqueness of AtELF3. Molecular Biology and Evolution, 41(10). https://doi.org/10.1093/molbev/msae205


